Background
I know it is very un-YouTube like to make a response video 4 years later, but I've only had the channel for a year so fight me.
However, I do have a very distinct memory of watching the video when it first came out, scrolling down to the comments and reading all the machine metaphor comments and thinking to myself: "Is that really what's going on??"
Four years later after fermenting in the back of my brain here we are.
Further reading
The absolute must-read source for more on this is Dan Nicholson's paper Is the cell really a machine? Like I said in the video, it has got to be mandatory reading for all biologists. If you want a companion to walk you through the paper, I'd reccommend Yogi Jaeger's video interpreting it in this lecture:
Of course, you should also probably watch some of the 'molecular machine' animations that are all over YouTube. Here are a few examples, including the ones I referenced in the video (Drew Berry's TED Talk and the Veritasium video).
Sources
A few years ago, Veritasium posted a video called "Your Body's Molecular Machines." The video showcases some amazing animations of the process of cell division, created by Drew Berry at the Walter and Eliza Hall Institute. But despite being super cool to look at, the animations are misleading.
Sources:
They make it seem as though our cells are no more than molecular clockwork and that at the bottom, we're just made out of nanomachines.
And if you scroll down to the comments, that's exactly the reaction you get: "dogs are just extra complex ps4s," "biology is just nanotechnology," "life is just the organisation of non-living components."
Sources:
Drew talks about 'molecular clockwork' at 8:00 in his TED talk and Derek mentions the parallels between our cellular 'machinery' and nanomachines at the 5:30 mark in his video.
But this machine metaphor is hardly true. It fails to capture the true nature of proteins, oversimplifies the cell, and gives us a false sense of confidence in how much we know about the biological world.
Sources:


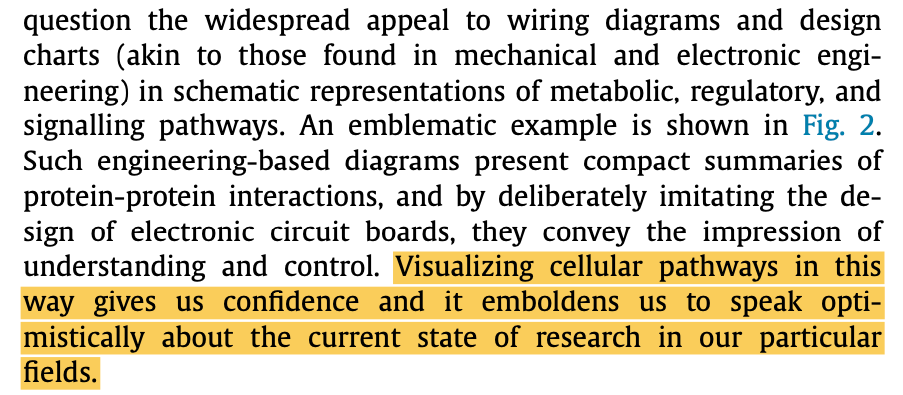
Don't get me wrong, Veritasium is one of my favourite channels. Derek is half the reason I've been inspired enough to make YouTube videos. Plus these animations would be an incredibly useful learning resource for students learning these processes for the first time.
But it's one thing for a high school student to use machine thinking as a learning tool, and a completely different thing to use the metaphor at the core of cancer biology. But we'll come back to that.
Source:

Let's go from the start. Why isn't the cell run by molecular machines like the animations suggest? Well, first we need to come up with some criteria to distinguish machines from non-machines. So what links together: trains, cars, computers, toasters, clocks and bikes?
Well, each of these machines is made up of smaller parts, which have relatively fixed, solid structures, and each part has its own specific function, like allowing the bike to move smoothly or telling you what hour it is.
Molecular machines seem to have all of these features too. At least, that's what it looks like in the animations. The molecular machine needed to copy your DNA is made up of smaller solid protein parts, and each of them is only doing one specific thing like splitting apart DNA into its two strands.
So this really is a machine. Except reality is a whole lot more complex than this.
Sources:
I imagine I'll get some pushback on my definition of a machine. I didn't want to turn the video into a definitional debate so I glossed over it pretty quickly. That was intentional.
Yes this is an overly-inclusive definition, including things we'd probably categorise as tools (e.g. a shovel) but then again, they would certainly be simple machines.
I also know that many machines have bendy parts, and that some machines have multi-functional parts. And yes, maybe in the future the line between machine and organism will be blurred. This is Joshua Bongard and Michael Levin's and perspective:

However, even as the title of the above paper points out, living things are certainly not 20th century machines (e.g. steam engines/computers in 2022) even if that changes in the future. I think the definition works fine for 'classical' machines. You can also read Dan's explanation for why functional specificity and structural rigidity are things we tend to focus on when thinking about protein complexes as 'molecular machines':
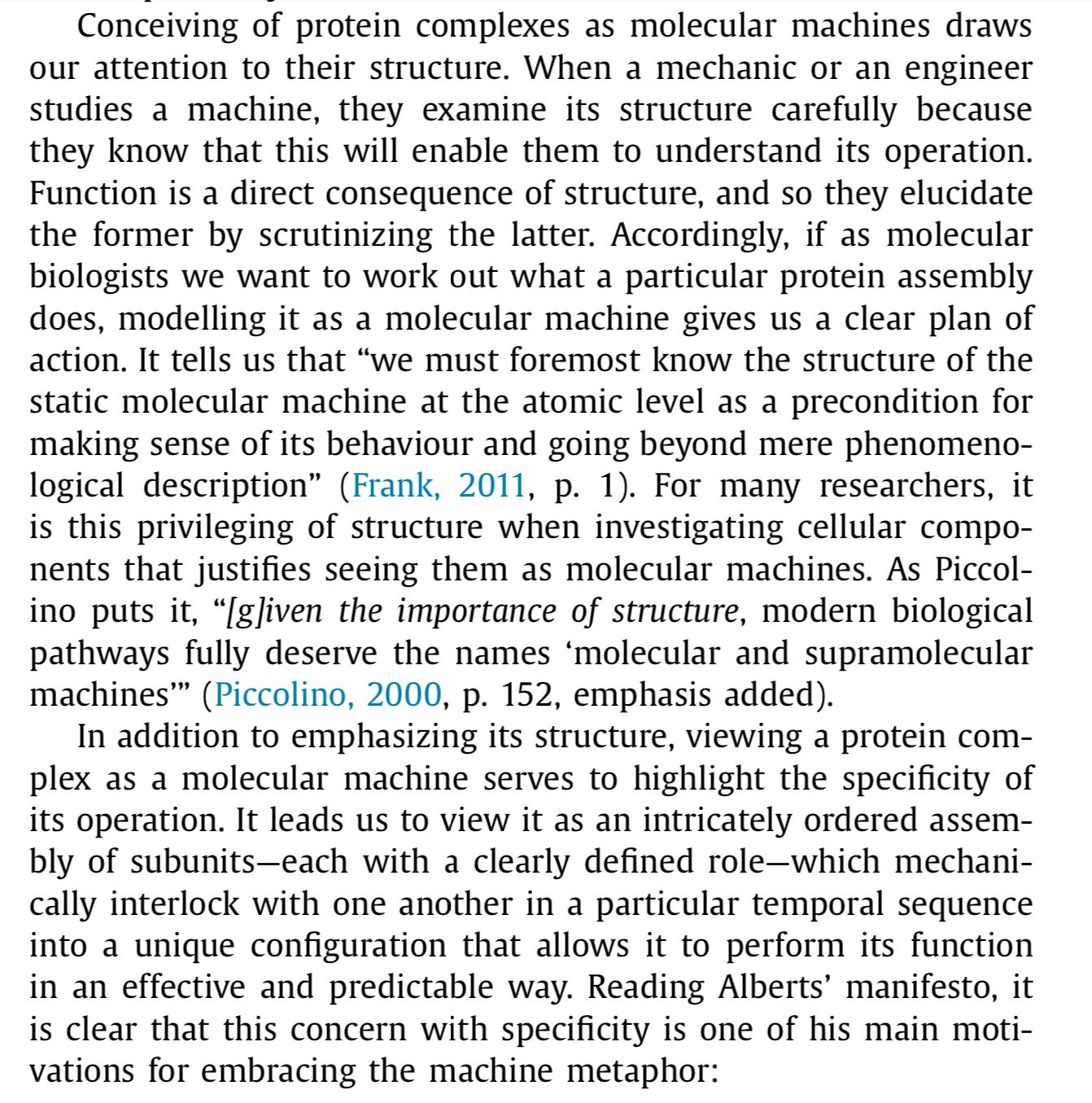
For starters, the structure of proteins isn't hard and rigid like we see in the animations. They're actually more like "dense liquids" that constantly jiggle around inside the cell.
Drew actually adds this jiggling into some of the later animations, but we can see it better in this simulation from the Skidmore Computational Biophysics Lab, led by Aurelia Ball.
Sources:
This is already strike one for the machine metaphor because, if my bike jiggled this much I wouldn't be able to go very far.
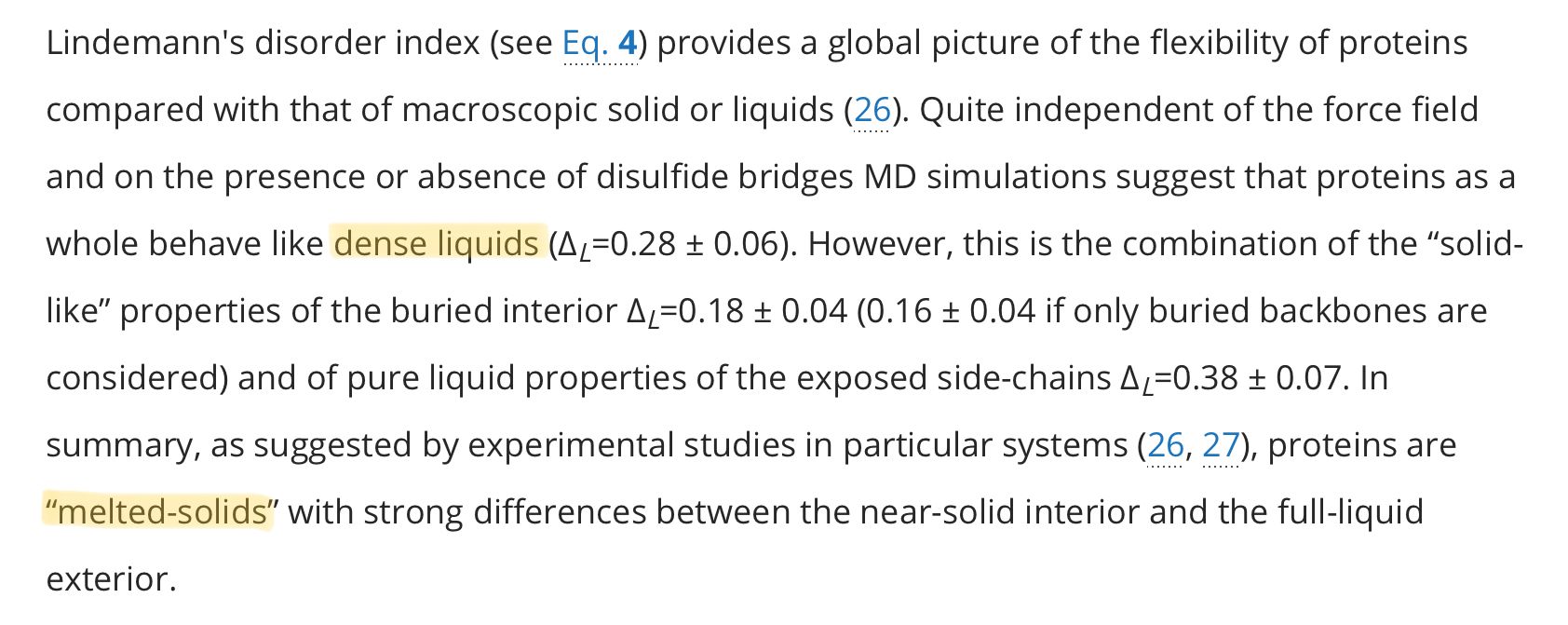
Normally, to analyse the structure of proteins, biochemists crystallise purified protein samples into a solid form first and then blast x-rays at them through a process known as X-ray crystallography. This has been the gold standard method for many years.
Source:

But if this is all we use, then it would be easy to fall into the trap of thinking all proteins are static, because you have to physically stop the protein from moving before you do anything. If your only tool is a hammer, you're going to think everything is a nail.
Source:
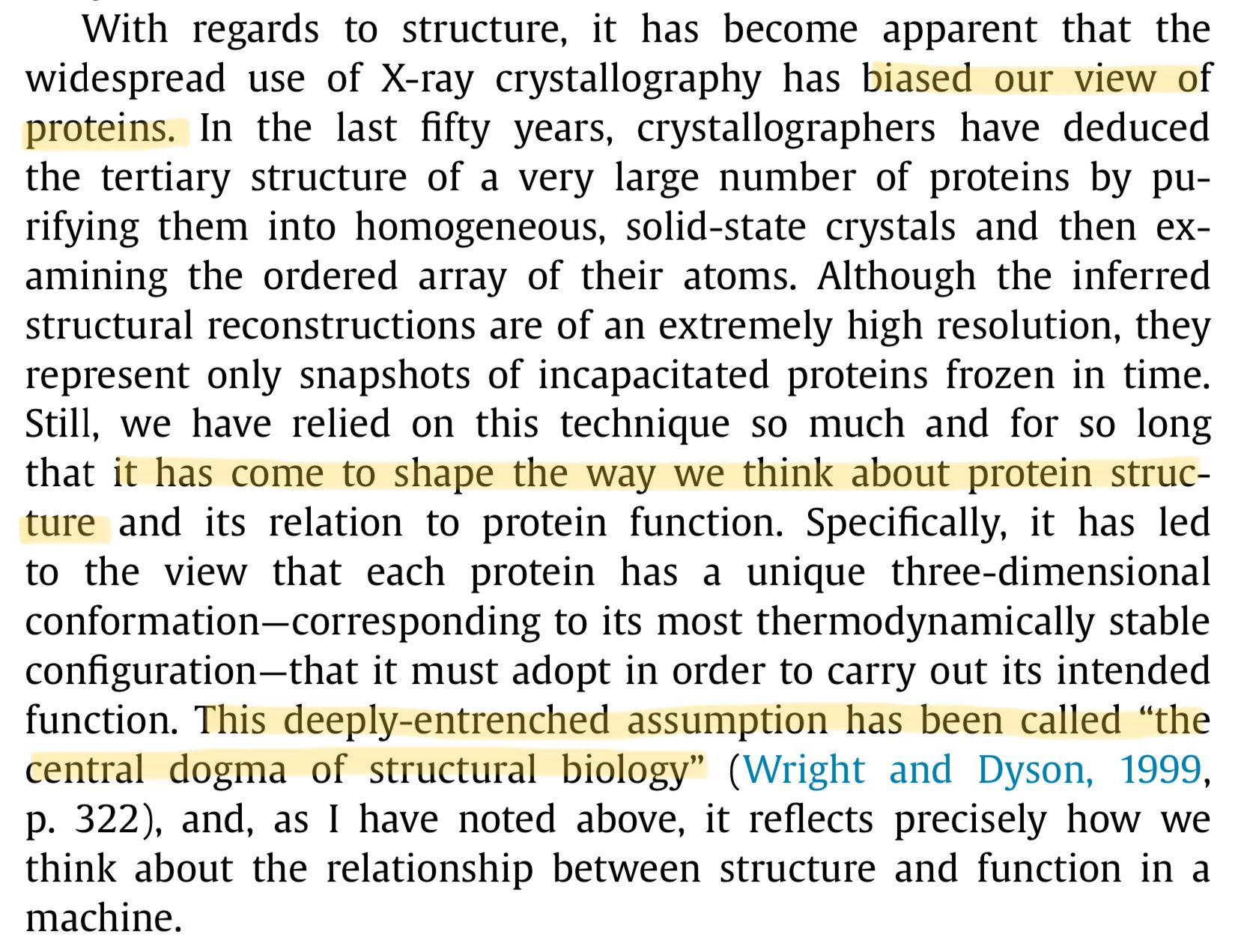
There are other methods which keep the protein jiggling in solution when it's being analysed and these results have been pretty revealing. Far from simple cogs in a machine, proteins have flexible, dynamic personalities. They open and close, twist around and can even rearrange into completely different structures, which brings us to point number 2.
Sources:
The most useful of these alternative tools is NMR spectroscopy which is now being used in tandem with X-ray crystallography:

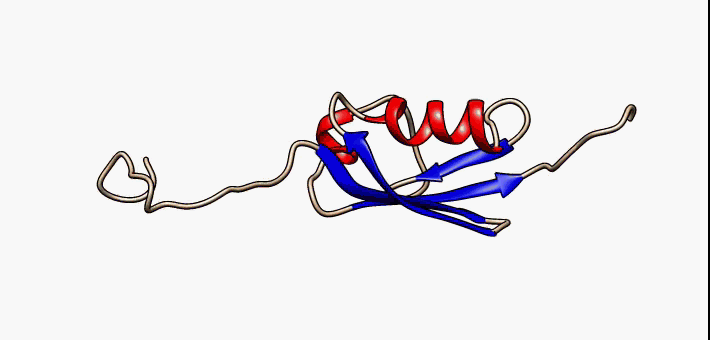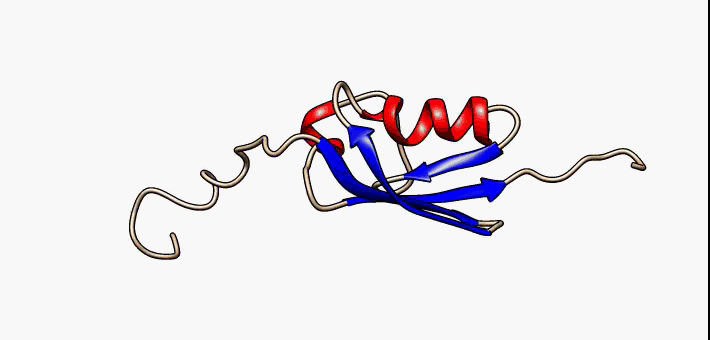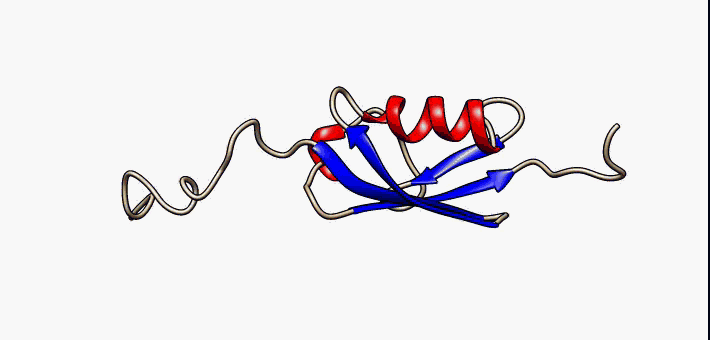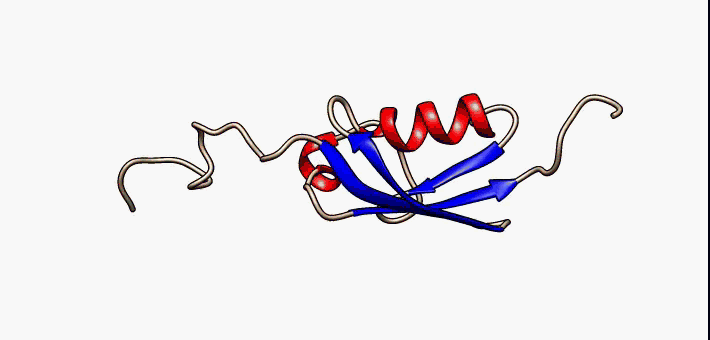
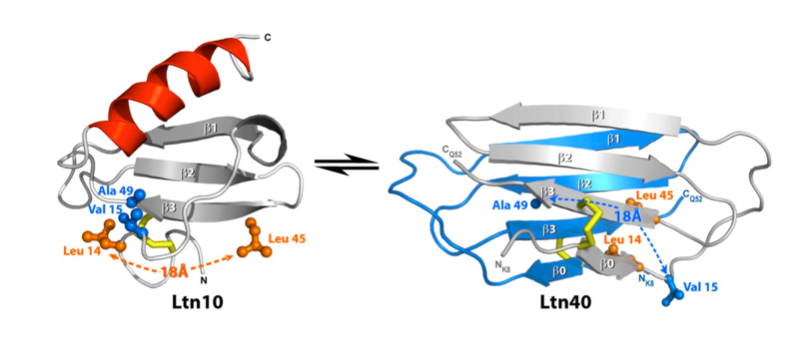

For many years, the reigning hypothesis has been that every protein has a single shape or conformation known as its native structure. This is also a myth. Proteins almost never have one shape, they have a bunch of different configurations that they shift between.
Source:
In one environment they might take on one shape, and then when they bind to a molecule or when the chemistry of the cell changes slightly, they might take on another shape. Proteins can even switch between conformations in the same conditions, so there’s at least some intrinsic randomness involved in protein structure.
Sources:

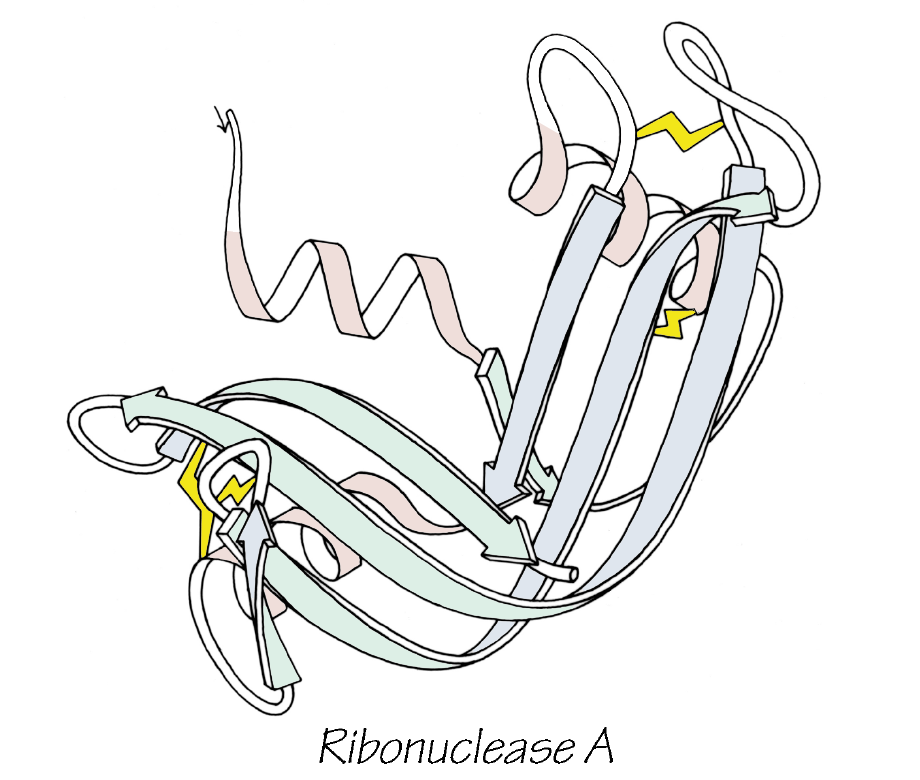
The extreme version of this are a group known as inherently disordered proteins or IDPs, and in mammals, they make up around a quarter of all proteins. IDPs are much more like "cooked spaghetti" than gears in a machine, they flop around with no set structure to them at all.
Source:


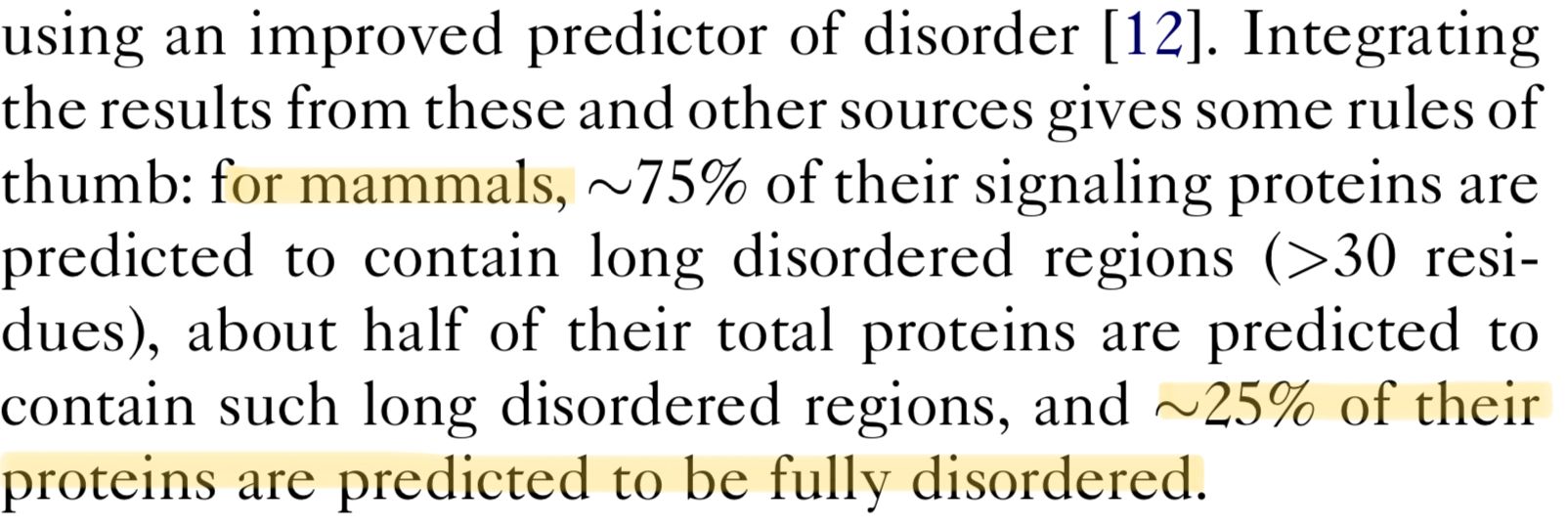
And this isn't an evolutionary mistake or anything either - disorder is often essential for the function of these proteins, because it allows them to bind to multiple different molecules. This flexibility just isn't possible with a single rigid structure.
Source:
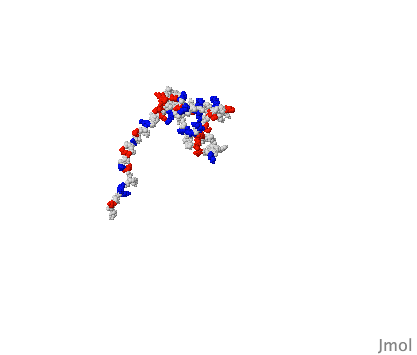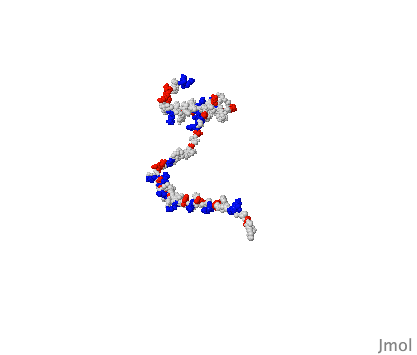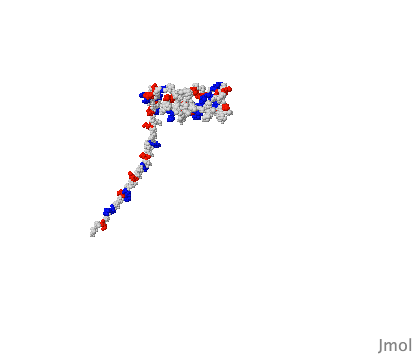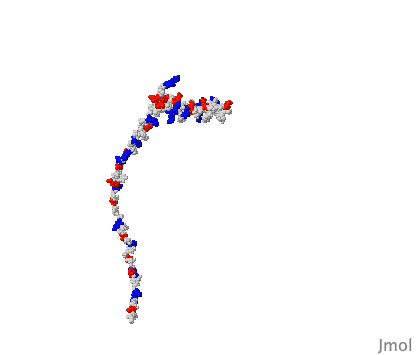
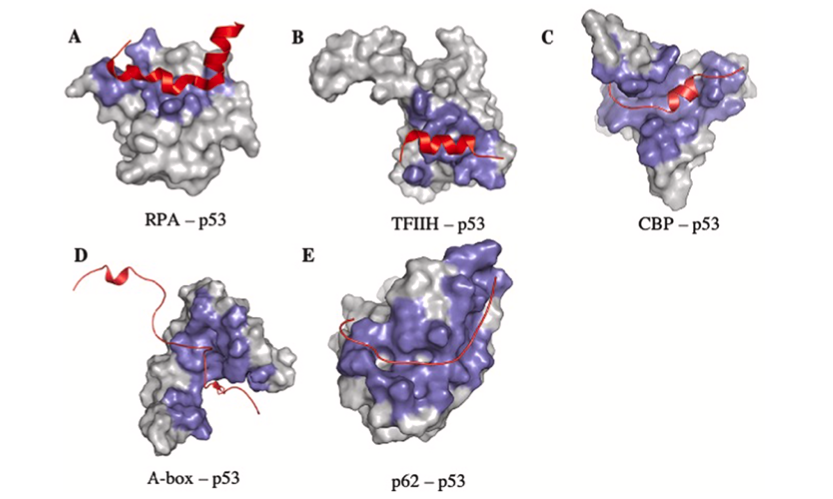

So the idea that an artificial intelligence like Google's AlphaFold has predicted the structure of every protein known to science is questionable because of the existence of these IDPs. I'm sure they’re are laughing at the AI trying to box their structure into one singular shape. As redditor sdreal wrote: "You can take a blurry picture of a person but you have no idea how well they can dance."
Sources:
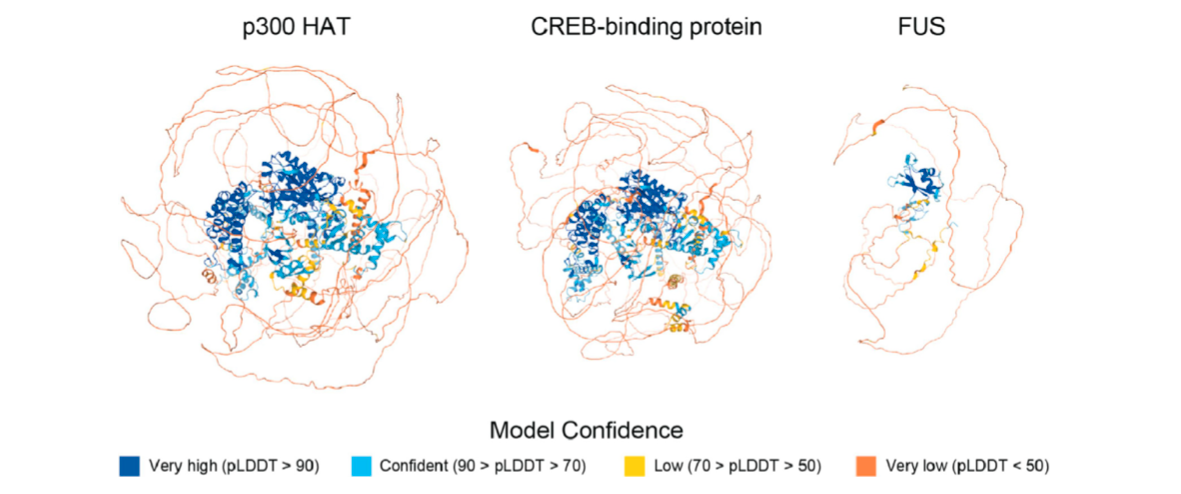
X-ray crystallography and computational techniques like AlphaFold are both still useful techniques for generating high resolution images of proteins. But we should realise that these tools are just the beginning for understanding protein structure. Looking at proteins through time is crucial for a complete understanding. We need to see how proteins dance.
Sources:
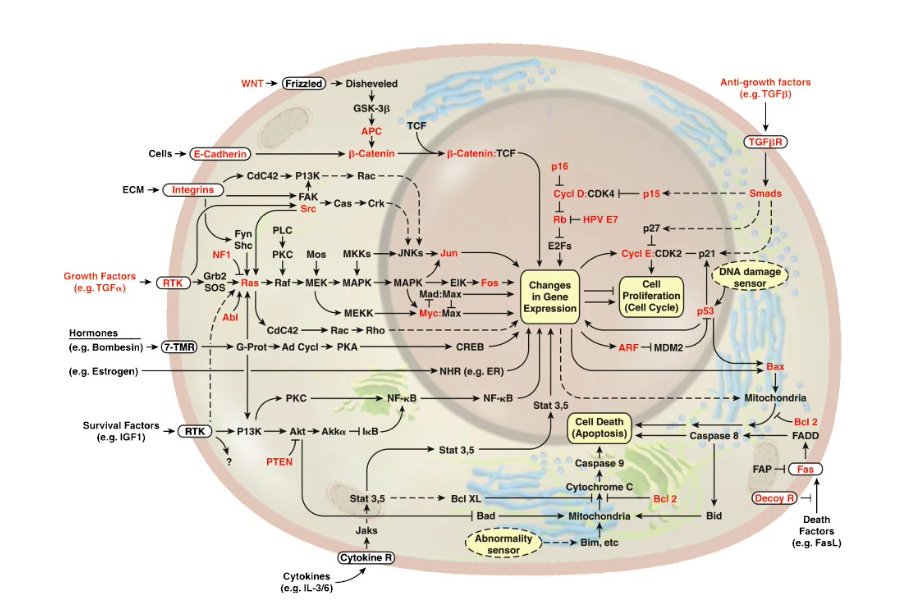
All in all, the physical structure of proteins is very different to that of machines, but surely we can still say that proteins have specific functions like the parts of machines, right? That's definitely the idea we get from these kinds of diagrams.
Sources:
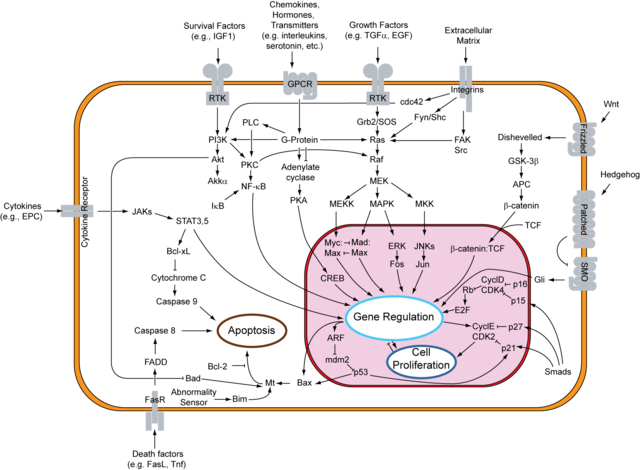
This is supposed to be a general map of some of the metabolic processes happening inside us. The design of diagrams like this is stolen from electrical engineering where each component in a circuit plays a clear function in the workings of the whole machine.
Source:
Is the analogy valid? No.
For many years, enzymes - a subgroup of proteins - have been thought to only interact with specific molecules. Take the enzyme methane monooxygenase or MMO. Like the name suggests, MMO was originally thought to convert only methane into methanol via a reaction known as hydroxylation.
Sources:
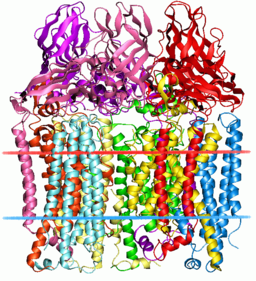
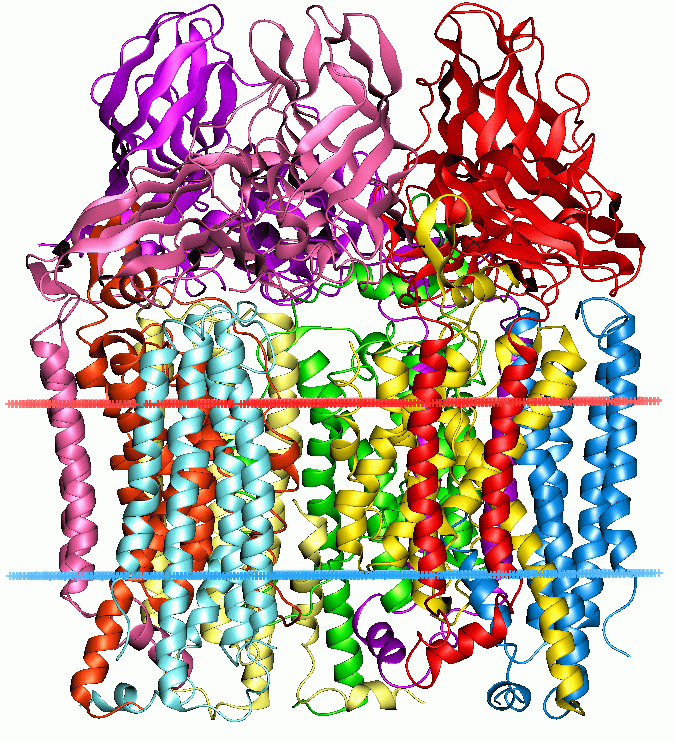
But we now know that in addition to methane, it can hydroxylate 150 other molecules. Biologists call this 'moonlighting' because in addition to its day job hydroxylating methane, MMO also works 150 other odd jobs around the cell to make ends meet.
Sources:

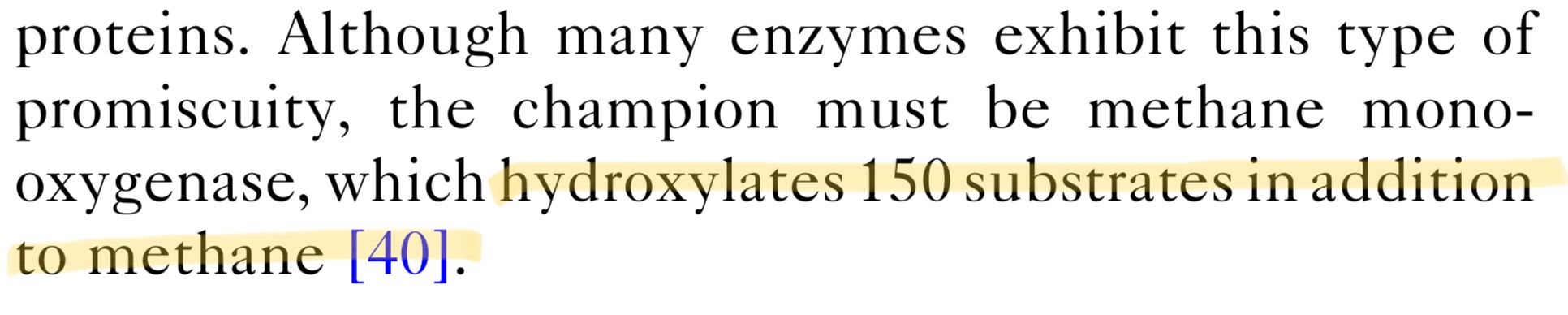
This is quite an extreme example - but the same thing occurs throughout the whole cell.
Source:

For instance, the ribosome works its day job turning RNA into protein. And actually, because this process looks so much like a conveyor belt, it’s become one of the most famous 'molecular machines'.
Source:
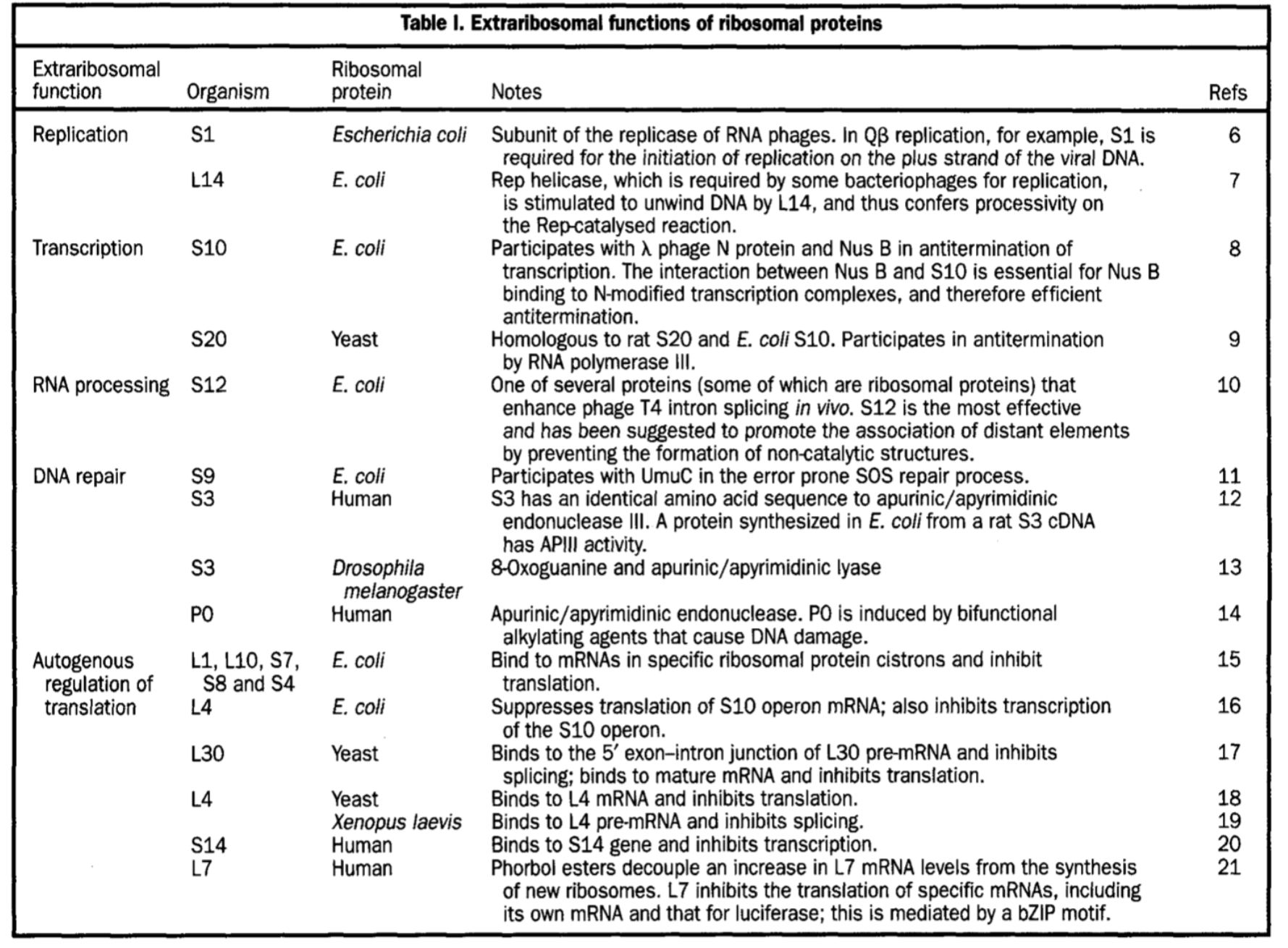
But we know now that it it too moonlights in other parts of the cell like in the nucleus replicating and repairing DNA. Often these moonlighting gigs are very hard to predict and they’re highly context dependent.
Source:
Technically I should have said parts of the ribosome (i.e. ribosomal proteins) moonlight in other areas, not the whole ribosome itself.
[EDIT: 17/10/22]
Charles Burridge (@CsBurridge) has fairly critiqued my point here, and I have responded as follows:
Evolution of the ribosome:
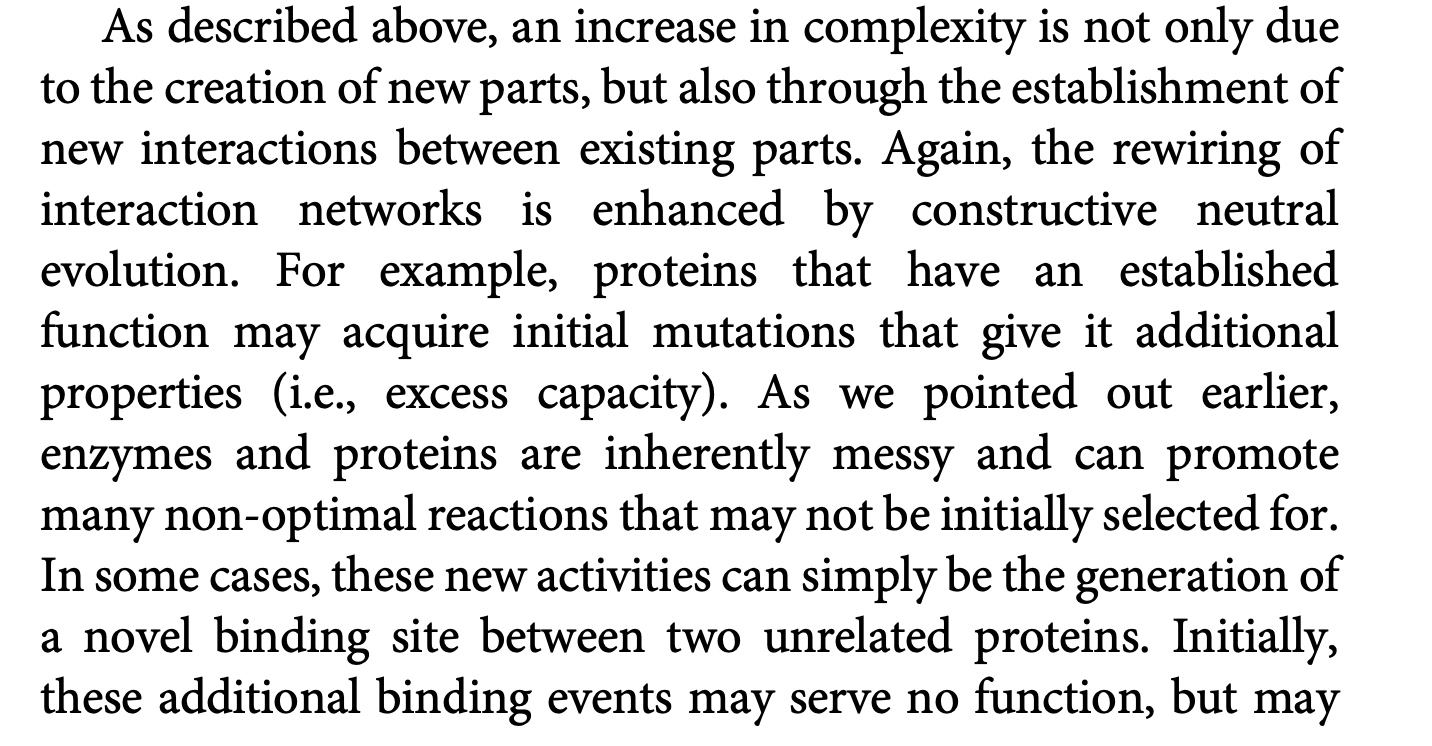
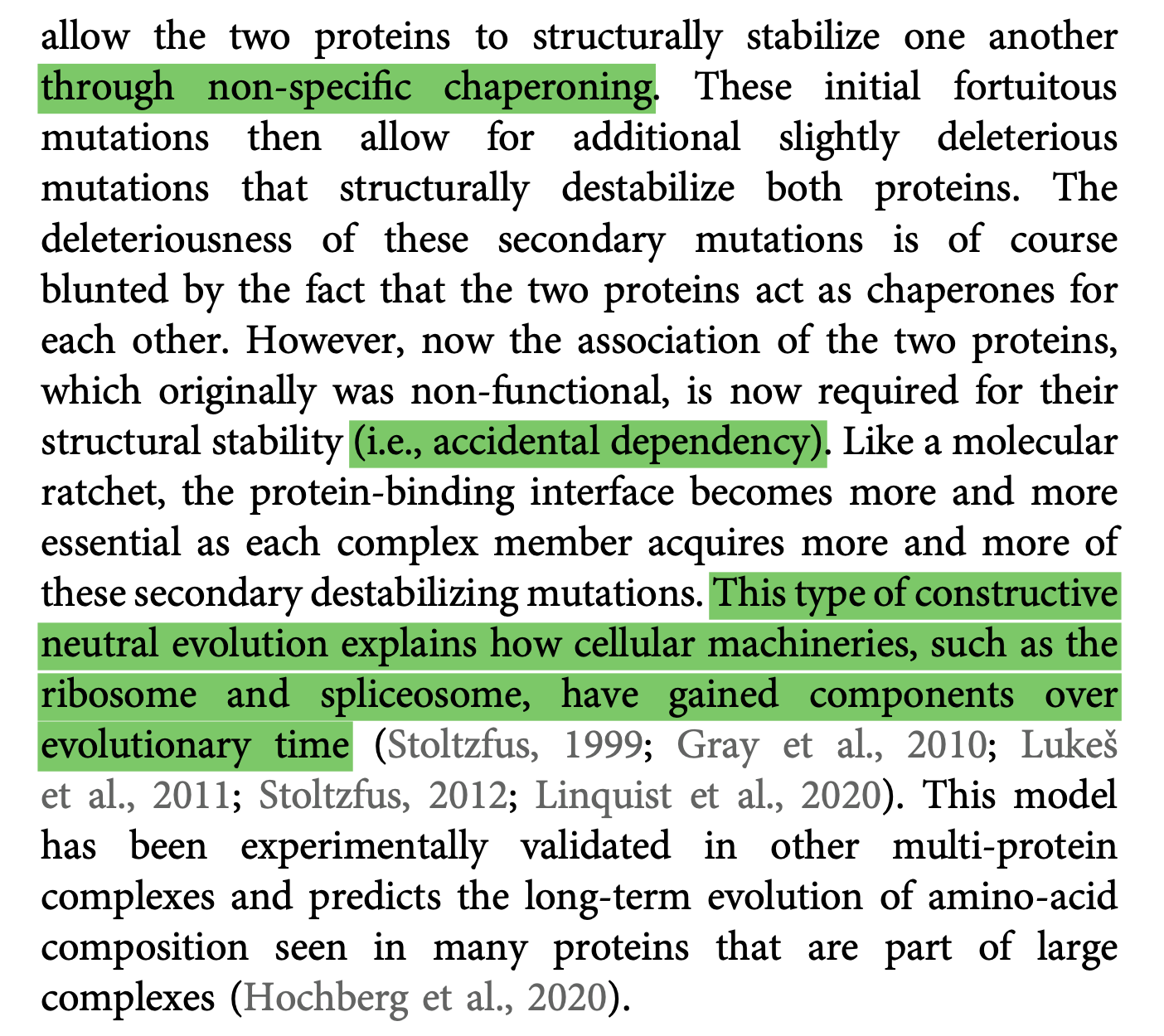
Dan Nicholson writes:
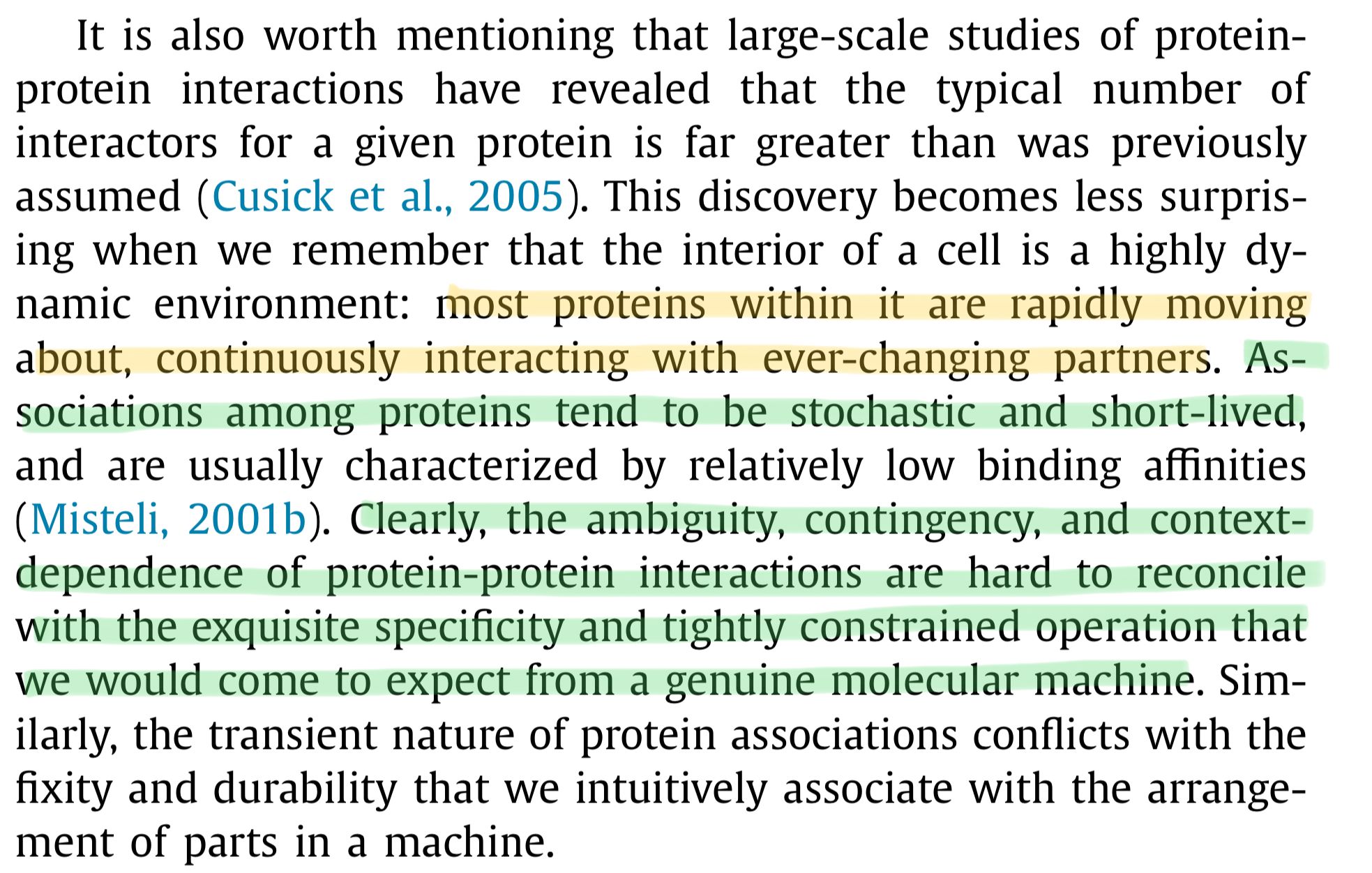
A protein can have very different functions, ... depending on where it is located in the cell, on the cell type in which it is expressed, [and] on the nature and number of proteins it binds to … most proteins within [the cell] are rapidly moving about, continuously interacting with ever-changing partners.


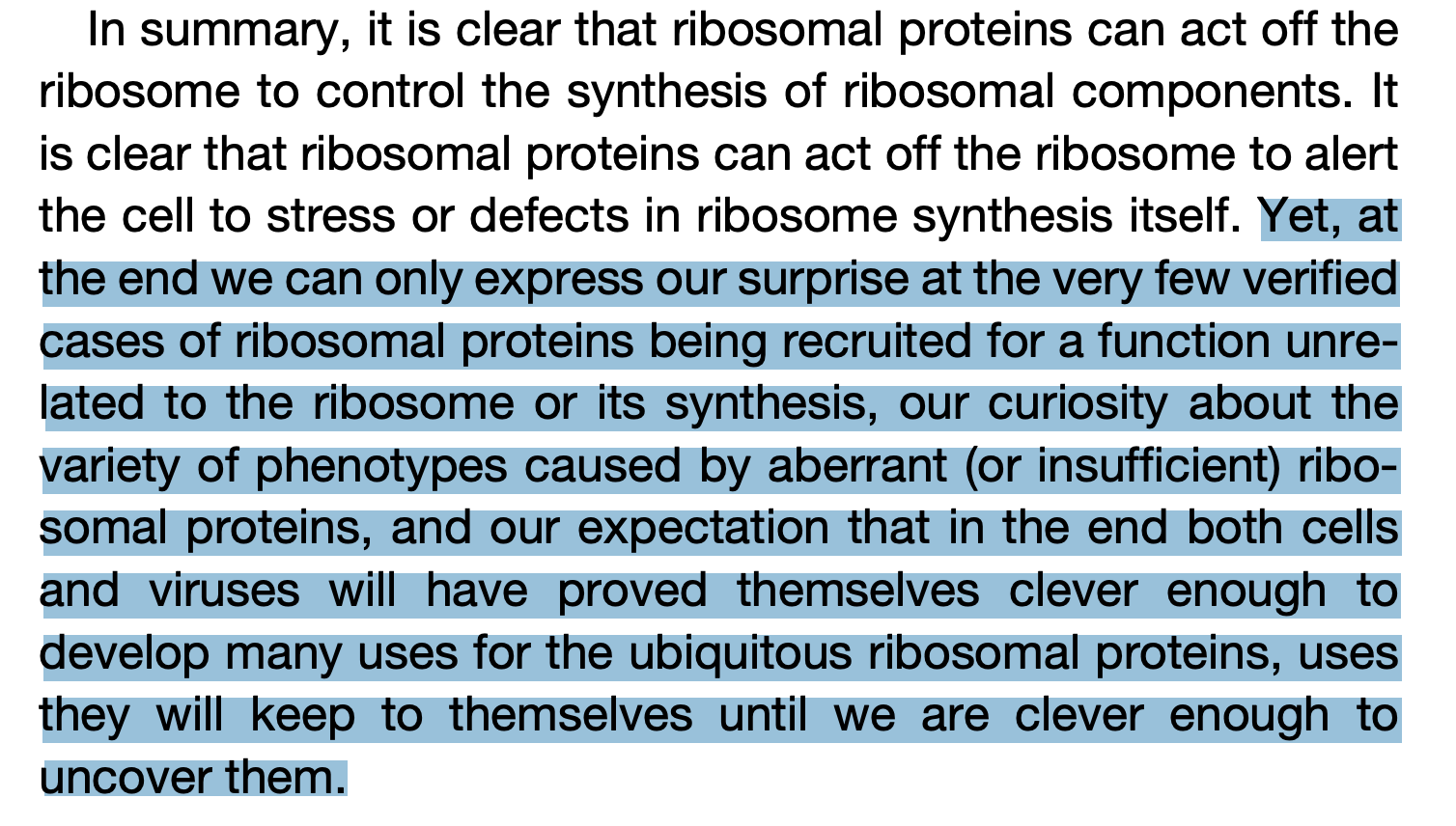
Pretty strange for parts of your machine to be doing a bunch of unpredictable jobs.
So because of this promiscuity, it’s a huge oversimplification to draw diagrams like this where proteins only have singular functions. These are only going to capture one possibility out of an essentially infinite number of ways proteins could interact.
Source:
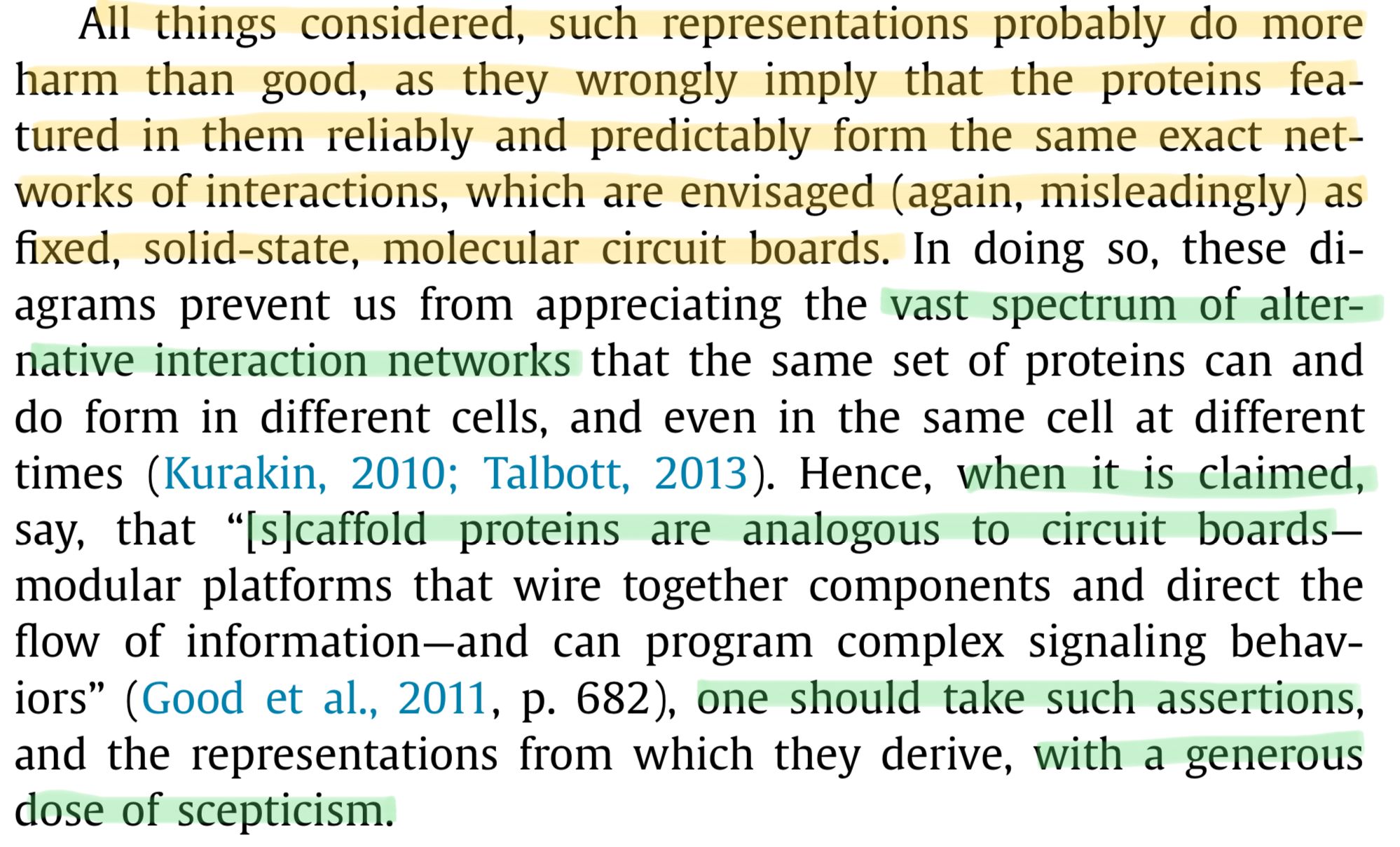
Again, quoting Dan Nicholson:
All things considered, such representations probably do more harm than good, as they wrongly imply that the proteins featured in them reliably and predictably form the same exact networks of interactions, which are envisaged (again, misleadingly) as fixed, solid-state, molecular circuit boards.
Source:
So why do we use these diagrams and these animations if they’re so misleading?
One commenter on the Veritasium video put it like this:
I like when humans finally caught up to this advanced biological process and had to compare it to machines that humans created that led to this discovery just to understand it.
Source:
Instead of accepting the complexity of life, we’ve reduced the cell to a machine to be more optimistic about how much we really know.
Here's an example of this in action.
One of the most influential papers in cancer biology published in 2000 was the "Hallmarks of cancer" by Douglas Hanahan and Robert Weinberg. It outlined six of the main capabilities of cancer and laid out a rough program for studying the disease ointo the 21st century. To date, it has over 39,000 citations which, in academia, is officially known as a shitton.
Source:
Hanahan, D., & Weinberg, R. A. (2000). The hallmarks of cancer. cell, 100(1), 57-70. https://doi.org/10.1016/S0092-8674(00)81683-9
It was so successful that they released a sequel in 2011 which has over 62,000 citations - also known as a metric shitton.
Source:
Hanahan, D., & Weinberg, R. A. (2011). Hallmarks of cancer: the next generation. cell, 144(5), 646-674. https://doi.org/10.1016/j.cell.2011.02.013
But at the heart of both papers is the machine metaphor and the idea that if we just map out all the functions of proteins in one ginormous map, we'll just have to run some maths and we'll know everything we need to know to cure cancer. In 2000 they wrote:
Two decades from now, having fully charted the wiring diagrams of every cellular signalling pathway, it will be possible to lay out the complete ‘integrated circuit of the cell.’
Source:
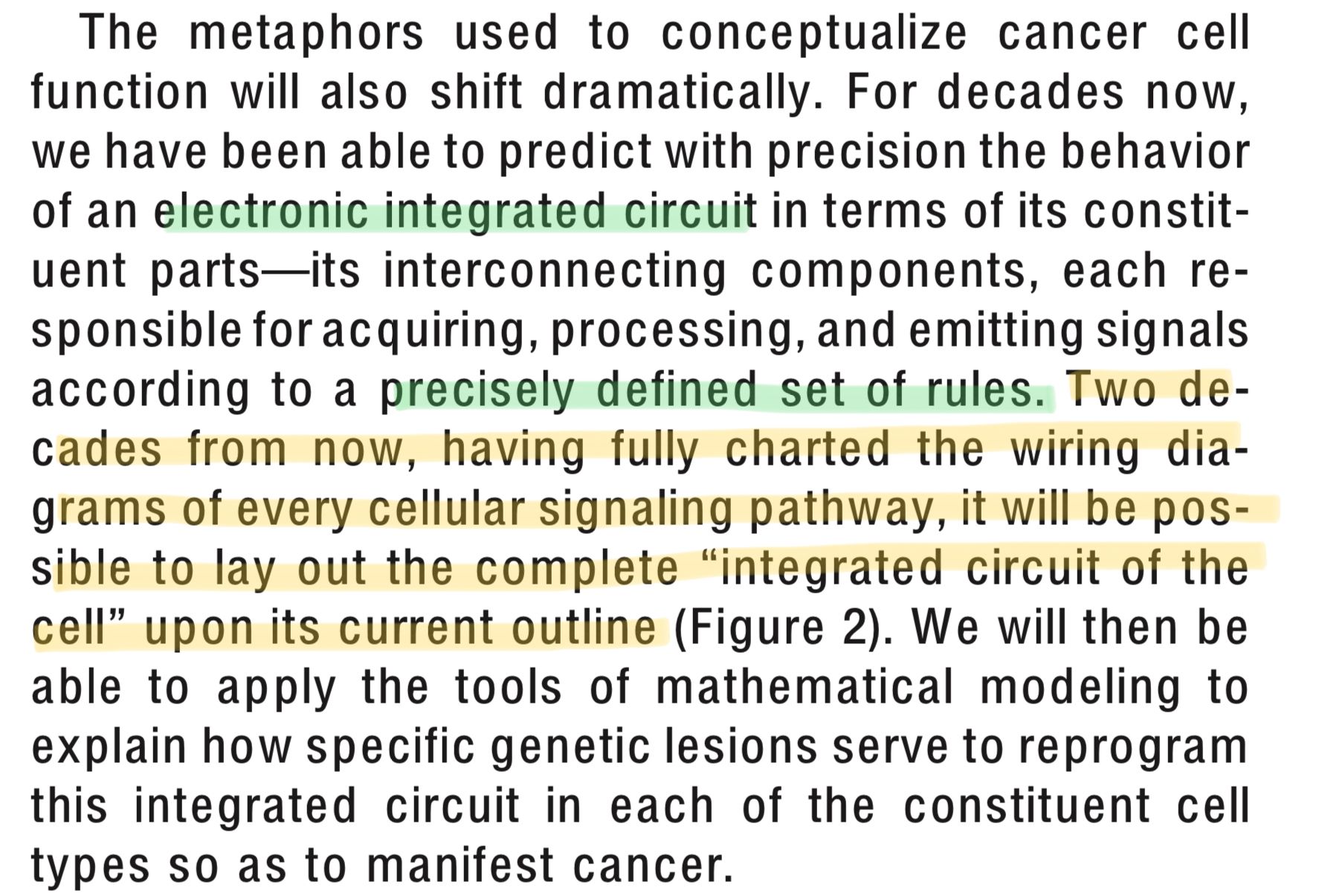
And two decades later, we've not even come close to 'fully charting the wiring diagrams' of the cell. Not because we haven't done enough science, but because it’s quite literally impossible thanks to the promiscuous nature of protein interactions.
Funnily enough, in the third instalment of the trilogy, published just this year, no circuitboard diagrams appear and the machine metaphor plays a much more minor role in their thinking.
Source:
Hanahan, D. (2022). Hallmarks of cancer: new dimensions. Cancer discovery, 12(1), 31-46. https://doi.org/10.1158/2159-8290.CD-21-1059
It is finally dawning on us that the cell is not a machine.
The metaphor has been useful to us, no doubt, but we've outgrown it and it's time we appreciated life in all of its complexity. Then, and only then, can we begin to understand the true nature of the cell.
Is it going to be much trickier than we initially thought? Of course, but who said science was easy?





Member discussion: